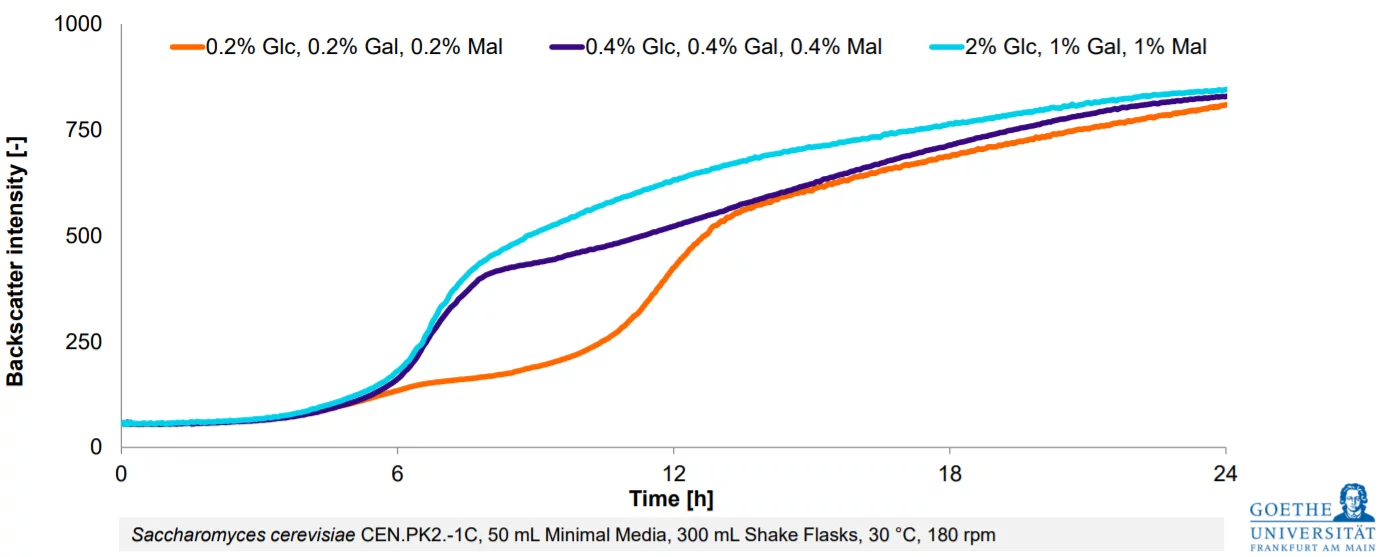
Data Spotlight
Monitoring Metabolic Events Through the Detection of eGFP Expressed by E. coli
Background
Inducing eGFP (enhanced Green Fluorescent Protein) expression in E. coli is a widely used method in molecular biology to study gene expression, protein localization, and other cellular processes.
The process begins by cultivating E. coli cells transformed with a plasmid containing the eGFP gene under the control of the inducible promoter. Once the culture reaches the desired growth phase—often during mid-log phase to maximize cellular activity—IPTG is added to initiate transcription of the eGFP gene. The timing and concentration of IPTG are critical, as over induction can lead to metabolic burden or inclusion body formation, affecting protein yield and functionality.
After induction, cells are monitored for fluorescence, or eGFP expression. The use of real-time monitoring tools or sampling for offline fluorescence measurements provides insights into the dynamics of protein production. This allows researchers to fine-tune experimental conditions, such as inducer concentration, temperature, and duration of expression, to maximize eGFP yield and quality.
Results
.jpg?width=900&name=Data%20Spotlight%20-%20%20Monitoring%20Metabolic%20Events%20Through%20the%20Detection%20of%20eGFP%20Expressed%20by%20E.%20coli%20(1).jpg)
Materials & Methods
A 250 mL shake flask was used for cultivation, filled with 10 mL of LB medium and inoculated with E.coli cells. The culture was shaken at 200 rpm/25 mm at 37°C.
The cultivation was monitored with the Multiparameter Sensor (MPS) which simultaneously monitored biomass (backscatter units, a.u.) and green fluorescence intensity at an emission of 555 nm. Dissolved oxygen levels were measured with the DO Sensor Pill. Real-time data was visualized and processed with the DOTS Software.
Conclusion
Real-time monitoring of multiple key parameters provides valuable insights into the dynamic metabolic processes occurring within a shake flask culture. In this study, the early IPTG inoculation, marked by the red triangle, induces protein expression, clearly demonstrating that GFP expression is closely coupled to the growth phase. Continuous tracking of these parameters allows researchers to better understand the interplay between growth and metabolic events, optimize experimental conditions, and identify critical timepoints for achieving more efficient and reproducible protein expression workflows.
The simplicity and visual nature of eGFP expression make it a powerful tool for both research and teaching, offering a clear, quantitative way to study gene regulation and optimize recombinant protein production in E. coli.
Have questions about your application?
Let’s work together to find a solution that works best for you.
From Estimation To High-Resolution Growth Curves


Customer Success Stories
.png)
-Kitana Manivone Kaiphanliam (Washington State University)
